Click on thumbnails to view full-size image.
Some are large files to show the details of these models; please be
patient while it downloads.
2009-2010

2008-2009
|
|
|
|
|
|
|
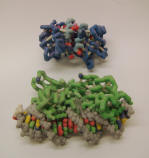
PhoB regulatory and effector domains with target DNA |
|
ZCorp Plaster Model |
|
|
|
|
|

|
|
|
|
PhoB regulatory domain, inactive and active forms |
|
ZCorp Plaster Model |
|
|
|
|
|
|

|
|
|
|
PhoB regulatory domain, dimerized active form |
|
ZCorp Plaster Model |
|
|
|
|
|

|
|
|
|
PhoB effector domain with target DNA |
|
ZCorp Plaster Model |
|
|
|
|
|
|

|
|
|
|
MtrA, regulatory and effector domains |
|
ZCorp Plaster Model |
|
|
|
|
|

|
|
|
|
MtrA, regulatory and effector domains, separated |
|
ZCorp Plaster Model |
|
|
|
|
|
|
2007-2008
|
|
|

|
|
|
|
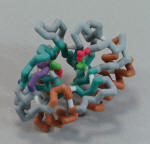
PlyB Lysin-Spacefill model |
|
ZCorp Plaster Model |
|
|
|
|
|
|
|
|
|
PlyB Lysin-Backbone model |
|
ZCorp Plaster Model |
|
|
|
|
|
|

|
|
|
|
Epimerase-Substrate in active site |
|
ZCorp Plaster Model |
|
|
|
|
|

|
|
|
|
Epimerase-No substrate in active site |
|
ZCorp Plaster Model |
|
|
|
|
|
|
|
|
More models from past years
|
|
|
|
|
|
|
|
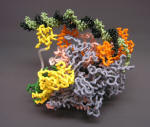
RNA Polymerase Holoenzyme-Open Promoter Complex |
|
Nylon Model |
|
|
|
|
|
|
|
|
|
|
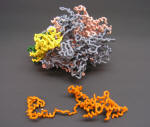
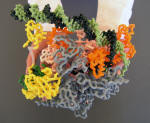
RNA Polymerase Core Enzyme with Sigma removed |
|
Nylon Model |
|
|
|
|
|
|
|
|
|
|
|
RNA Polymerase Holoenzyme |
|
Nylon Model |
|
|
|
|
|
|

|
|
|
|
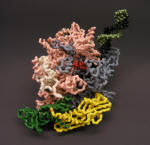
RNA Polymerase Holoenzyme: separated to expose active site and to remove nucleic acids/sigma factor |
|
Nylon Model |
|
|
|
|
|
|
|
|
|
|
|
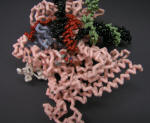
RNA Polymerase Ternary Elongation Complex |
|
RNA transcript colored red. Nylon Model |
|
|
|
|
|
|
|
|
|
|
RNA Polymerase Ternary Elongation Complex |
|
Active site amino acids (seven conserved residues, NADFDGD) shown with sidechains. Mg2+ ion colored purple. Nylon Model |
|
|
|
|
|
|
|

|
|
|
|
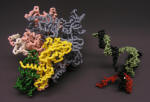
RNA Polymerase Ternary Elongation Complex: model separated to expose active site |
|
Nylon Model |
|
|
|
|
|
|
|
|
|
|
RNA Polymerase Ternary Elongation Complex: Nucleic acids removed from RNAP core enzyme |
|
Nylon Model |
|
|
|
|
|
|
|
|
|
|
|
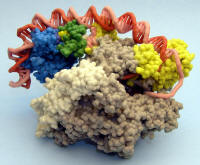
Prokaryotic RNA Polymerase Activation Complex |
|
ZCorp plaster model. Part of beta subunit removed to expose transcription bubble. |
|
|
|
|
|
|
|
|
|
|
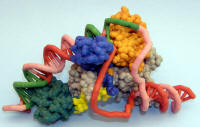
Sigma factor (promoter recognition subunit) complexed with promoter DNA |
|
ZCorp plaster model. Colors on sigma show various domains. Grey is part of core RNAP. Green on DNA are promoter regions. |
|
|
|
|
|
|
|
|
|
|
|
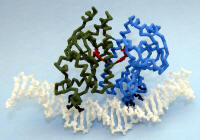
Cyclic AMP Receptor Protein (CRP) or Catabolite Activator Protein (CAP) |
|
Nylon model. Cyclic AMP colored Red. Dimer interacts with UP-element (region upstream of the promoter DNA) and plays a role in increasing transcription levels. |
|
|
|
|
|
|
|
|
|
|
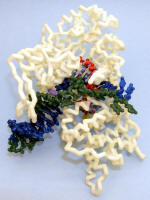
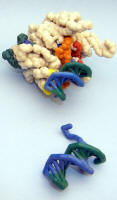
T7 phage RNA Polymerase elongation complex |
|
Nylon model. Based on PDB ID: 1MSW. |
|
|
|
|
|
|
|
|
|
|
|
T7 phage RNA Polymerase Initiation (Promoter) complex |
|
ZCorp plaster model. Based on PDB ID: 1CEZ. |
|
|
|
|
|
|

|
|
|
|
T7 phage RNA Polymerase elongation complex |
|
ZCorp plaster model. Based on PDB ID: 1MSW. |
|
|
|
|
|
|